Edited, memorised or added to reading queue
on 17-Feb-2020 (Mon)
Do you want BuboFlash to help you learning these things? Click here to log in or create user.
Flashcard 5005069651212
| status | not learned | measured difficulty | 37% [default] | last interval [days] | |||
|---|---|---|---|---|---|---|---|
| repetition number in this series | 0 | memorised on | scheduled repetition | ||||
| scheduled repetition interval | last repetition or drill |
Open it
When several links are connected together by joints, they are said to form a kinematic chain. Links containing only two joint elements are called binary links, those having three joint elements are called ternary links, those having four joint elements are called quaternary lin
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Getting started with Merge Requests | GitLab
s. When you create a new feature branch, change the files, and push it to GitLab, you have the option to create a Merge Request, which is essentially a request to merge one branch into another. <span>The branch you added your changes into is called source branch while the branch you request to merge your changes into is called target branch. The target branch can be the default or any other branch, depending on the branching strategies you choose. In a merge request, beyond visualizing the differences between the original c
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
"Work In Progress" merge requests | GitLab
the “Work In Progress” flag to a merge request Removing the “Work In Progress” flag from a merge request Including/excluding WIP merge requests when searching “Work In Progress” merge requests <span>If a merge request is not yet ready to be merged, perhaps due to continued development or open threads, you can prevent it from being accepted before it’s ready by flagging it as a Work In Progress. This will disable the “Merge” button, preventing it from being merged, and it will stay disabled until the “WIP” flag has been removed. Adding the “Work In Progress” flag to a merge request There are several ways to flag a merge request as a Work In Progress: Add [WIP] or WIP: to the start of the merge request’s title.
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
[1904.09378v1] A General Neural Network Architecture for Persistence Diagrams and Graph Classification
Journal reference ACM classification MSC classification Report number arXiv identifier DOI ORCID arXiv author ID Help pages Full text Search Download PDF Statistics > Machine Learning Title:<span>A General Neural Network Architecture for Persistence Diagrams and Graph Classification Authors:Mathieu Carrière , Frédéric Chazal , Yuichi Ike , Théo Lacombe , Martin Royer , Yuhei Umeda (Submitted on 20 Apr 2019 (this version), latest version 17 Oct 2019 (v3 )) Abstract: Graph classification is a difficult problem that has drawn a lot of attention from the machine learning community over the past few years. This is mainly due to the fact that, contrarily to Euclidean vectors, the inherent complexity of graph structures can be quite hard to encode and handle for traditional classifiers. Even though kernels have been proposed in the literature, the increase in the dataset sizes has greatly limited the use of kernel methods since computation and storage of kernel matrices has become impracticable. In this article, we propose to use extended persistence diagrams to efficiently encode graph structure. More precisely, we show that using the so-called heat kernel signatures for the computation of these extended persistence diagrams allows one to quickly and efficiently summarize the graph structure. Then, we build on the recent development of neural networks for point clouds to define an architecture for (extended) persistence diagrams which is modular and easy-to-use. Finally, we demonstrate the usefulness of our approach by validating our architecture on several graph datasets, on which the obtained results are comparable to the state-of-the-art for graph classification. Subjects: Machine Learning (stat.ML); Computational Geometry (cs.CG); Machine Learning (cs.LG); Algebraic Topology (math.AT) Cite as: arXiv:1904.09378 [stat.ML] (or arXiv:1904.09378v1 [
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
[1905.04579v2] Are Powerful Graph Neural Nets Necessary? A Dissection on Graph Classification
al reference ACM classification MSC classification Report number arXiv identifier DOI ORCID arXiv author ID Help pages Full text Search Download PDF Computer Science > Machine Learning Title:<span>Are Powerful Graph Neural Nets Necessary? A Dissection on Graph Classification Authors:Ting Chen , Song Bian , Yizhou Sun (Submitted on 11 May 2019 (v1 ), last revised 24 May 2019 (this version, v2)) Abstract: Graph Neural Nets (GNNs) have received increasing attentions, partially due to their superior performance in many node and graph classification tasks. However, there is a lack of understanding on what they are learning and how sophisticated the learned graph functions are. In this work, we first propose Graph Feature Network (GFN), a simple lightweight neural net defined on a set of graph augmented features. We then propose a dissection of GNNs on graph classification into two parts: 1) the graph filtering, where graph-based neighbor aggregations are performed, and 2) the set function, where a set of hidden node features are composed for prediction. We prove that GFN can be derived by linearizing graph filtering part of GNNs, and leverage it to test the importance of the two parts separately. Empirically we perform evaluations on common graph classification benchmarks. To our surprise, we find that, despite the simplification, GFN could match or exceed the best accuracies produced by recently proposed GNNs, with a fraction of computation cost. Our results suggest that linear graph filtering with non-linear set function is powerful enough, and common graph classification benchmarks seem inadequate for testing advanced GNN variants. Comments: A shorter version titled "Graph Feature Networks" was accepted to ICLR'19 RLGM workshop Subjects: Machine Learning (cs.LG); Social and Information Networks (cs.SI); Machine Learning (stat.ML) Cite as: arXiv:1905.04579 [cs.LG] (or arXiv:1905.04579v2 [cs.LG] for this version
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
[1904.08745v2] edGNN: a Simple and Powerful GNN for Directed Labeled Graphs
al reference ACM classification MSC classification Report number arXiv identifier DOI ORCID arXiv author ID Help pages Full text Search Download PDF Computer Science > Machine Learning Title:<span>edGNN: a Simple and Powerful GNN for Directed Labeled Graphs Authors:Guillaume Jaume , An-phi Nguyen , María Rodríguez Martínez , Jean-Philippe Thiran , Maria Gabrani (Submitted on 18 Apr 2019 (v1 ), last revised 4 Dec 2019 (this version, v2)) Abstract: The ability of a graph neural network (GNN) to leverage both the graph topology and graph labels is fundamental to building discriminative node and graph embeddings. Building on previous work, we theoretically show that edGNN, our model for directed labeled graphs, is as powerful as the Weisfeiler-Lehman algorithm for graph isomorphism. Our experiments support our theoretical findings, confirming that graph neural networks can be used effectively for inference problems on directed graphs with both node and edge labels. Code available at this https URL . Comments: Representation Learning on Graphs and Manifolds @ ICLR19 Subjects: Machine Learning (cs.LG); Machine Learning (stat.ML) Cite as: arXiv:1904.08745 [cs.LG] (or arXiv:1904.08745v2 [cs.LG] for this version) Submission history From: An-Phi Nguyen
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Publications | MIT | Caroline Uhler
ink] PUBLICATIONS [emptylink] [emptylink] [emptylink] [emptylink] GROUP [emptylink] [emptylink] [emptylink] [emptylink] TEACHING [emptylink] [emptylink] [emptylink] [emptylink] CONFERENCES More <span>Preprints Ordering-based causal structure learning in the presence of latent variables D. I. Bernstein, B. Saeed, C. Squires and C. Uhler Under review [arXiv ] Keywords: causal inference, algebraic statistics Permutation-based causal structure learning with unknown intervention targets C. Squires, Y. Wang and C. Uhler Under review [arXiv ] Keywords: causal inference, applications to biology Overparameterized neural networks can implement associative memory A. Radhakrishnan, M. Belkin and C. Uhler Under review [arXiv ] Keywords: autoencoders, machine learning Covariance matrix estimation under total positivity for portfolio selection R. Agrawal, U. Roy and C. Uhler Under review [arXiv ] Keywords: total positivity, applications to finance Learning high-dimensional Gaussian graphical models under total positivity without tuning parameters Y. Wang, U. Roy and C. Uhler Under review [arXiv ] Keywords: total positivity, applications to finance Anchored causal inference in the presence of measurement error B. Saeed, A. Belyaeva, Y. Wang and C. Uhler Under review [arXiv ] Keywords: causal inference, applications to biology Total positivity in structured binary distributions S. Lauritzen, C. Uhler and P. Zwiernik Under review [arXiv ] Keywords: total positivity, algebraic statistics Brownian motion tree models are toric B. Sturmfels, C. Uhler and P. Zwiernik Under review [arXiv ] Keywords: total positivity, algebraic statistics Autoencoder and optimal transport to infer single-cell trajectories of biological processes K.D. Yang, K. Damodaran, S. Venkatchalapathy, A.C. Soylemezoglu, G.V Shivashankar and C. Uhler Under review [arXiv ] Keywords: autoencoders, application to biology Maximum likelihood estimation for totally positive log-concave densities E. Robeva, B. Sturmfels, N. Tran and C. Uhler Under review [arXiv ] Keywords: shape-constrained density estimation, total positivity High-dimensional joint estimation of multiple directed Gaussian graphical models Y. Wang, S. Segarra and C. Uhler Under review [arXiv ] Keywords: causal inference, high-dimensional statistics Consistency guarantees for permutation-based causal inference algorithms L. Solus, Y. Wang, L. Matejovicova and C. Uhler Under review [arXiv ] Keywords: causal inference, algebraic statistics 2019 Algebraic statistics in practice: Applications to networks M. Casanellas, S. Petrovic and C. Uhler To appear in Annual Review of Statistics and its Applications (invited review)
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Publications | MIT | Caroline Uhler
nsistency guarantees for permutation-based causal inference algorithms L. Solus, Y. Wang, L. Matejovicova and C. Uhler Under review [arXiv ] Keywords: causal inference, algebraic statistics <span>2019 Algebraic statistics in practice: Applications to networks M. Casanellas, S. Petrovic and C. Uhler To appear in Annual Review of Statistics and its Applications (invited review) [arXiv ] Keywords: causal inference, algebraic statistics Maximum likelihood estimation in Gaussian models under total positivity S. Lauritzen, C. Uhler and P. Zwiernik Annals of Statistics 47 (2019), pp. 1835-1863. [arXiv ] Keywords: total positivity, Gaussian graphical models Multi-domain translation by learning uncoupled autoencoders K.D. Yang and C. Uhler Computational Biology Workshop, International Conference on Machine Learning (ICML 2019) [arXiv ] Keywords: autoencoders, machine learning Memorization in overparameterized autoencoders A. Radhakrishnan, K.D. Yang, M. Belkin and C. Uhler Deep Phenomena Workshop, International Conference on Machine Learning (ICML 2019) [arXiv ] Keywords: autoencoders, machine learning ABCD-Strategy: Budgeted experimental design for targeted causal structure discovery R. Agrawal, C. Squires, K.D. Yang, K. Shanmugam and C. Uhler Proceedings of Machine Learning Research 89 (AISTATS 2019), pp. 3400-3409 Keywords: causal inference, experimental design Size of interventional Markov equivalence classes in random DAG models D. Katz-Rogozhnikov,K. Shanmugam, C. Squires and C. Uhler Proceedings of Machine Learning Research 89 (AISTATS 2019), pp. 3234-3243 Keywords: causal inference Scalable unbalanced optimal transport using generative adversarial networks K.D. Yang and C. Uhler International Conference on Learning Representations (ICLR 2019) [arXiv ] Keywords: optimal transport, applications to biology Geometry of discrete copulas E. Perrone, L. Solus and C. Uhler Journal of Multivariate Analsysis 172 (2019), pp. 162-179 [arXiv ] Keywords: algebraic statistics Loading monotonicity of weighted premiums, and total positivity properties of weight functions D. Richards and C. Uhler Journal of Mathematical Analysis and Applications, 475 (2019), pp. 532-553. [arXiv ] Keywords: mathematical statistics, total positivity Generalized Fréchet bounds for cell entries in multidimensional contingency tables C. Uhler and D. Richards Journal of Algebraic Statistics 10 (special issue for Stephen E. Fienberg) (2019), pp. 1-12 [arXiv ] Keywords: mathematical statistics, total positivity Geometry of log-concave density estimation E. Robeva, B. Sturmfels and C. Uhler Discrete & Computational Geometry 61 (2019), pp. 136-160 [arXiv ] Keywords: shape-constrained density estimation, algebraic statistics 2018 Direct estimation of differences in causal graphs Y. Wang, C. Squires, A. Belyaeva and C. Uhler Advances in Neural Information Processing Systems 31 (2018) [arXiv ] Keywords: causal inference, applications to biology PatchNet: Interpretable neural networks for image classification A. Radhakrishnan, C. Durham, A. Soylemezoglu and C. Uhler Machine Learning for Health (ML4H) Workshop, Neural Information Processing Systems (2018) [arXiv ] Keywords: neural nets, applications to biology Minimal I-MAP MCMC for scalable structure discovery in causal DAG models R. Agrawal, T. Broderick and C. Uhler Proceedings of Machine Learning Research 80 (ICML 2018), pp. 89-98 [arXiv ] Keywords: causal inference, Bayesian statistics, applications to biology Characterizing and learning equivalence classes of causal DAGs under interventions K. D. Yang, A. Katcoff and C. Uhler Proceedings of Machine Learning Research 80 (ICML 2018), pp. 5537-5546 [arXiv ] Keywords: causal inference, applications to biology Learning directed acyclic graphs based on sparsest permutations G. Raskutti and C. Uhler Stat 7 (2018), e183 [arXiv ] Keywords: causal inference, algebraic statistics Counting Markov equivalence classes for DAG models on trees A. Radhakrishnan, L. Solus and C. Uhler Discrete Applied Mathematics 244 (2018), pp. 170-185 [arXiv ] Keywords: causal inference, algebraic statistics Nuclear mechanopathology and cancer diagnosis C. Uhler and G.V. Shivashankar Trends in Cancer 4 (2018), pp. 320-331 (invited review) [journal ] Keywords: applications to biology, chromosome packing Generalized permutohedra from probabilistic graphical models F. Mohammadi, C. Uhler, C. Wang and J. Yu SIAM Journal on Discrete Mathematics 32 (2018), pp. 64-93 [arXiv ] Keywords: causal inference, algebraic statistics Exact formulas for the normalizing constants of Wishart distributions for graphical models C. Uhler, A. Lenkoski and D. Richards Annals of Statistics 46 (2018), pp. 90-118 [arXiv ] Keywords: Gaussian graphical models, Bayesian statistics Gaussian graphical models: An algebraic and geometric perspective C. Uhler Chapter for Handbook on Graphical Models (editors: M. Drton, S. Lauritzen, M. Maathuis and M. Wainwright) [arXiv ] Keywords: Gaussian graphical models, algebraic statistics 2017 Machine learning for nuclear mechano-morphometric biomarkers in cancer diagnosis A. Radhakrishnan, D. Damodaran, A. Soylemezoglu, C. Uhler, and G.V. Shivashankar Scientific Repor
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Publications | MIT | Caroline Uhler
pective C. Uhler Chapter for Handbook on Graphical Models (editors: M. Drton, S. Lauritzen, M. Maathuis and M. Wainwright) [arXiv ] Keywords: Gaussian graphical models, algebraic statistics <span>2017 Machine learning for nuclear mechano-morphometric biomarkers in cancer diagnosis A. Radhakrishnan, D. Damodaran, A. Soylemezoglu, C. Uhler, and G.V. Shivashankar Scientific Reports 7 (2017), article nr. 17946. [journal ] Keywords: neural nets, applications to biology Permutation-based causal inference algorithms with interventions Y. Wang, L. Solus, K.D. Yang and C. Uhler Advances in Neural Information Processing (NIPS 2017) [arXiv ] Keywords: causal inference, algebraic statistics Network analysis identifies chromosome intermingling regions as regulatory hotspots for transcription A. Belyaeva, S. Venkatachalapathy, M. Nagarajan, G.V. Shivashankar and C. Uhler Proceedings of the National Academy of Sciences, U.S.A. 114 (2017), pp. 13714-13719. [journal ] Keywords: chromosome packing, applications to biology Regulation of genome organization and gene expression by nuclear mechanotransduction C. Uhler and G.V. Shivashankar Nature Reviews Molecular Cell Biology 18 (2017), pp. 717-727 (invited review). [journal ] Keywords: applications to biology, chromosome packing Chromosome intermingling: Mechanical hotspots for genome regulation C. Uhler and G.V. Shivashankar Trends in Cell Biology 27 (2017), pp. 810-819 (invited review) [journal ] Keywords: chromosome packing, applications to biology Orientation and repositioning of chromosomes correlate with cell geometry-dependent gene expression Y. Wang, M. Nagarajan, C. Uhler and G.V. Shivashankar Molecular Biology of the Cell 28 (2017), pp. 1997-2009 [journal ] Keywords: chromosome packing, applications to biology Maximum likelihood estimation for linear Gaussian covariance models P. Zwiernik, C. Uhler and D. Richards Journal of the Royal Statistical Society, Series B 79 (2017), pp. 1269-1292 [arXiv ] Keywords: Gaussian models, covariance estimation, optimization Counting Markov equivalence classes by number of immoralities A. Radhakrishnan, L. Solus and C. Uhler Proceedings of the Thirty-Third Conference on Uncertainty in Artificial Intelligence (UAI 2017) [arXiv ] Keywords: causal inference, algebraic statistics Total positivity in Markov structures S. Fallat, L. Lauritzen, K. Sadeghi, C. Uhler, N. Wermuth and P. Zwiernik Annals of Statistics 45 (2017), pp. 1152-1184 [arXiv ] Keywords: total positivity, mathematical statistics Exact goodness-of-fit testing for the Ising model A. Martin del Campo, S. Cepeda and C. Uhler Scandinavian Journal of Statistics 44 (2017), pp. 285-306 [arXiv ] Keywords: hypothesis testing, algebraic statistics 2016 Extremal positive semidefinite matrices for graphs without K5 minors L. Solus, C. Uhler and R. Yoshida Linear Algebra and its Applications 509 (2016), pp. 247-275 [arXiv ] Keywords: Gaussian graphical models, algebraic statistics Geometric control and modeling of genome reprogramming C. Uhler and G.V. Shivashankar BioArchitecture 6 (2016), pp. 76-84 [journal ] Keywords: chromosome packing, applications to biology Exponential varieties M. Michalek, B. Sturmfels, C. Uhler and P. Zwiernik Proceedings of the London Mathematical Society 112 (2016), pp. 27-56 [arXiv ] Keywords: Gaussian graphical models, algebraic statistics 2015 Faithfulness and learning of hypergraphs from discrete distributions A. Klimova, C. Uhler and T. Rudas Journal of Computational Statistics and Data Analysis 87 (2015), pp. 57-72. [arXiv ] Keywords: causal inference, algebraic statistics 2014 Hypersurfaces and their singularities in partial correlation testing S. Lin, C. Uhler, B. Sturmfels and P. Bühlmann Foundations of Computational Mathematics 14 (2014), pp. 1079-1116 [arXiv ] Keywords: causal inference, algebraic statistics Differentially private logistic regression for detecting multiple-SNP association in GWAS databases F. Yu, M. Rybar, C. Uhler and S.E. Fienberg Privacy in Statistical Databases 8744 (2014), pp. 170-184 [arXiv ] Keywords: differential privacy, applications to biology Sphere packing with limited overlap M. Iglesias-Ham, M. Kerber and C. Uhler Proceedings of the 26th Canadian Conference on Computational Geometry, Halifax, Nova Scotia (2014), pp. 155-161. [arXiv ] Keywords: sphere packing, applications to biology Scalable privacy-preserving data sharing methodology for genome-wide association studies F. Yu, S.E. Fienberg, A. Slavkovic and C. Uhler Journal of Biomedical Informatics 50 (2014), pp. 133-141 [arXiv ] Keywords: differential privacy, applications to biology 2013 Packing ellipsoids with overlap C. Uhler and S.J. Wright SIAM Review 55 (2013), pp. 671-706 (selected as Research Spotlight) [arXiv ] Keywords: optimization, sphere packing, chro
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Article 5016262151436
Levy-Kramer-2018-Reproducible_Data_Science
#has-images #machine-learning #software-engineering #unfinished
https://www.slideshare.net/joshlk100/reproducible-data-science-review-of-pachyderm-data-version-control-and-git-lfs-tools
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
machine learning - What are the pros and cons of using DVC and Pachyderm? - Stack Overflow
-control data-science dvc pachyderm share |improve this question asked Jul 4 '19 at 6:12 [emptylink] [imagelink] 020jfm01 020jfm01 1 add a comment | 1 Answer 1 active oldest votes 2 [emptylink] <span>It depends on what are you trying to accomplish. DVC will help you with organizing the ML experimentation process . Pachyderm is more focused on data engineering pipelines. share |improve this answer edited Jul 5 '19 at 19:11 answered Jul 5 '19 at 16:28 [emptylink] [imagelink] Mr. Outis Mr. Outis 5111 silver badge55 bronze badges add a comment | Your Answe
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
How does this compare to pachyderm? | Hacker News
Feb 10, 2019 | parent | favorite | on: DVC – Open Source Machine Learning Version Control... How does this compare to pachyderm? [emptylink] [emptylink] ishcheklein on Feb 10, 2019 [emptylink] <span>From a very high level perspective - Pachyderm is a data engineering tool designed with ML in mind, DVC is a tool to organize and version control an ML project. Probably, one way to think would be Spark/Hadoop or Airflow vs Git/Github. [emptylink] [emptylink] AlexCoventry on Feb 11, 2019 [emptylink] Thanks. Applications are open for YC Summer 2020 Guidelines | FAQ | Support | API | Security | Lists | Bookmarklet | Leg
Article 5016282860812
Sato_Wider_Windheuser-2019-Continuous_Delivery_Machine_Learning-martinfowler,com
#has-images #machine-learning #software-engineering #unfinished
Refactoring Agile Architecture About ThoughtWorks Continuous Delivery for Machine Learning Automating the end-to-end lifecycle of Machine Learning applications Machine Learning applications are becoming popular in our industry, however the process for developing, deploying, and continuously improving them is more complex compared to more traditional software, such as a web service or a mobile application. They are subject to change in three axis: the code itself, the model, and the data. Their behaviour is often complex and hard to predict, and they are harder to test, harder to explain, and harder to improve. Continuous Delivery for Machine Learning (CD4ML) is the discipline of bringing Continuous Delivery principles and practices to Machine Learning applications. 19 September 2019 Danilo Sato bio Arif Wider bio Christoph Windheuser bio CONTINUOUS DELIVERY BIG DATA CONTENTS Introduction and Definition A Machine L
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Article 5016310385932
2019-What_is_Kubernetes-kubernetes,io
#frameworks #has-images #software-engineering #unfinished
Documentation Blog Partners Community Case Studies English v1.17 Concepts HOME GETTING STARTED CONCEPTS TASKS TUTORIALS REFERENCE CONTRIBUTE Concepts Overview What is Kubernetes Kubernetes Components The Kubernetes API Working with Kubernetes Objects Understanding Kubernetes Objects Kubernetes Object Management Names Namespaces Labels and Selectors Annotations Field Selectors Recommended Labels Cluster Architecture Nodes Master-Node Communication Controllers Concepts Underlying the Cloud Controller Manager Containers Images Container Environment Variables Runtime Class Container Lifecycle Hooks Workloads Pods Pod Overview Pods Pod Lifecycle Init Containers Pod Preset Pod Topology Spread Constraints Disruptions Ephemeral Containers Controllers ReplicaSet ReplicationController Deployments
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
2019-What_is_Kubernetes-kubernetes,io
workloads and services, that facilitates both declarative configuration and automation. It has a large, rapidly growing ecosystem. Kubernetes services, support, and tools are widely available. <span>The name Kubernetes originates from Greek, meaning helmsman or pilot. Google open-sourced the Kubernetes project in 2014. Kubernetes builds upon a decade and a half of experience that Google has with running production workloads at scale , combined with b
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
2019-What_is_Kubernetes-kubernetes,io
on and automation. It has a large, rapidly growing ecosystem. Kubernetes services, support, and tools are widely available. The name Kubernetes originates from Greek, meaning helmsman or pilot. <span>Google open-sourced the Kubernetes project in 2014. Kubernetes builds upon a decade and a half of experience that Google has with running production workloads at scale , combined with best-of-breed ideas and practices from the community.
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
2019-What_is_Kubernetes-kubernetes,io
ng production workloads at scale , combined with best-of-breed ideas and practices from the community. Going back in time Let’s take a look at why Kubernetes is so useful by going back in time. <span>Traditional deployment era: Early on, organizations ran applications on physical servers. There was no way to define resource boundaries for applications in a physical server, and this caused resource allocation issues. For example, if multiple applications run on a physical server, there can be instances where one application would take up most of the resources, and as a result, the other applications
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
2019-What_is_Kubernetes-kubernetes,io
: Early on, organizations ran applications on physical servers. There was no way to define resource boundaries for applications in a physical server, and this caused resource allocation issues. <span>For example, if multiple applications run on a physical server, there can be instances where one application would take up most of the resources, and as a result, the other applications would underperform. A solution for this would be to run each application on a different physical server. But this did not scale as resources were underutilized, and it was expensive for organizations to maintain many physical servers. Virtualized deployment era: As a solution, virtualization was introduced. It allows you to run multiple Virtual Machines (VMs) on a single physical server’s CPU. Virtualization allows a
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
2019-What_is_Kubernetes-kubernetes,io
ould be to run each application on a different physical server. But this did not scale as resources were underutilized, and it was expensive for organizations to maintain many physical servers. <span>Virtualized deployment era: As a solution, virtualization was introduced. It allows you to run multiple Virtual Machines (VMs) on a single physical server’s CPU. Virtualization allows applications to be isolated between VMs and provides a level of security as the information of one application cannot be freely accessed by another application. Virtualization allows better utilization of resources in a physical server and allows better scalability because an application can be added or updated easily, reduces hardware costs, a
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
2019-What_is_Kubernetes-kubernetes,io
ver’s CPU. Virtualization allows applications to be isolated between VMs and provides a level of security as the information of one application cannot be freely accessed by another application. <span>Virtualization allows better utilization of resources in a physical server and allows better scalability because an application can be added or updated easily, reduces hardware costs, and much more. With virtualization you can present a set of physical resources as a cluster of disposable virtual machines. Each VM is a full machine running all the components, including its own operating system, on top of the virtualized hardware. Container deployment era: Containers are similar to VMs, bu
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
2019-What_is_Kubernetes-kubernetes,io
n application can be added or updated easily, reduces hardware costs, and much more. With virtualization you can present a set of physical resources as a cluster of disposable virtual machines. <span>Each VM is a full machine running all the components, including its own operating system, on top of the virtualized hardware. Container deployment era: Containers are similar to VMs, but they have relaxed isolation properties to share the Operating System (OS) among the applications. Therefore, containers are
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
2019-What_is_Kubernetes-kubernetes,io
of physical resources as a cluster of disposable virtual machines. Each VM is a full machine running all the components, including its own operating system, on top of the virtualized hardware. <span>Container deployment era: Containers are similar to VMs, but they have relaxed isolation properties to share the Operating System (OS) among the applications. Therefore, containers are considered lightweight. Similar to a VM, a container has its own filesystem, CPU, memory, process space, and more. As they are decoupled from the underlying infrastructure, they are portable across clouds and OS distributions. Containers have become popular because they provide extra benefits, such as: Agile application creation and deployment: increased ease and efficiency of container image creation compare
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
2019-What_is_Kubernetes-kubernetes,io
running on one big single-purpose machine. Resource isolation: predictable application performance. Resource utilization: high efficiency and density. Why you need Kubernetes and what can it do <span>Containers are a good way to bundle and run your applications. In a production environment, you need to manage the containers that run the applications and ensure that there is no downtime. For example, if a container goes down, another container needs to start. Wouldn’t it be easier if this behavior was handled by a system? That’s how Kubernetes comes to the rescue! Kubernetes provides you with a framework to run distributed systems resiliently. It takes care of scaling and failover for your application, provides deployment patterns, and more. For example, Kubernetes can easily manage a canary deployment for your system. Kubernetes provides you with: Service discovery and load balancing Kubernetes can expose a container usin
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
2019-What_is_Kubernetes-kubernetes,io
keys. You can deploy and update secrets and application configuration without rebuilding your container images, and without exposing secrets in your stack configuration. What Kubernetes is not <span>Kubernetes is not a traditional, all-inclusive PaaS (Platform as a Service) system. Since Kubernetes operates at the container level rather than at the hardware level, it provides some generally applicable features common to PaaS offerings, such as deployment, scaling, load balancing, logging, and monitoring. However, Kubernetes is not monolithic, and these default solutions are optional and pluggable. Kubernetes provides the building blocks for building developer platforms, but preserves user choice and flexibility where it is important. Kubernetes: Does not limit the types of applications supported. Kubernetes aims to support an extremely diverse variety of workloads, including stateless, stateful, and data-processing
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Bash Special Parameters Explained with 4 Example Shell Scripts
plained with 4 Example Shell Scripts [imagelink] ≡ Menu Home Free eBook Start Here Contact About Bash Special Parameters Explained with 4 Example Shell Scripts by Sasikala on May 12, 2010 Tweet <span>As part of our on-going bash tutorial series, we discussed about bash positional parameters in our previous article. In this article let us discuss about the bash special parameters with few practical shell script examples. Some of the bash special parameters that we will discuss in this article are: $*, $@, $#, $$, $!, $?, $-, $_ To access the whole list of positional parameters, the two special parameters $* and $@ are available. Outside of double quotes, these two are equivalent: Both expand to the list of positional parameters starting with $1 (separated by spaces). Within double quotes, however, they differ: $* within a pair of double quotes is equivalent to the list of positional parameters, separated by the first character of IFS “$1c$2c$3…”. $@ within a pair of double quotes is equivalent to the list of positional parameters, separated by unquoted spaces, i.e., “$1” “$2″..”$N”. Example 1: Use Bash $* and $@ to Expand Positional Parameters This example shows the value available in $* and $@. First, create the expan.sh as shown below. $ cat expan.sh #!/bin/bash export IFS='-' cnt=1 # Printing the data available in $* echo "Values of \"\$*\":" for arg in "$*" do echo "Arg #$cnt= $arg" let "cnt+=1" done cnt=1 # Printing the data available in $@ echo "Values of \"\$@\":" for arg in "$@" do echo "Arg #$cnt= $arg" let "cnt+=1" done Next, execute the expan.sh as shown below to see how $* and $@ works. $ ./expan.sh "This is" 2 3 Values of "$*": Arg #1= This is-2-3 Values of "$@": Arg #1= This is Arg #2= 2 Arg #3= 3 The above script exported the value of IFS (Internal Field Separator) with the ‘-‘. There are three parameter passed to the script expan.sh $1=”This is”,$2=”2″ and $3=”3″. When printing the each value of special parameter “$*”, it gives only one value which is the whole positional parameter delimited by IFS. Whereas “$@” gives you each parameter as a separate word. Example 2: Use $# to Count Positional Parameters $# is the special parameter in bash which gives you the number of positional parameter in decimal. First, create the arithmetic.sh as shown below. $ cat arithmetic.sh #!/bin/bash if [ $# -lt 2 ] then echo "Usage: $0 arg1 arg2" exit fi echo -e "\$1=$1" echo -e "\$2=$2" let add=$1+$2 let sub=$1-$2 let mul=$1*$2 let div=$1/$2 echo -e "Addition=$add\nSubtraction=$sub\nMultiplication=$mul\nDivision=$div\n" If the number of positional parameters is less than 2, it will throw the usage information as shown below, $ ./arithemetic.sh 10 Usage: ./arithemetic.sh arg1 arg2 Example 3: Process related Parameters – $$ and $! The special parameter $$ will give the process ID of the shell. $! gives you the process id of the most recently executed background process. The following script prints the process id of the shell and last execute background process ID. $ cat proc.sh #!/bin/bash echo -e "Process ID=$$" sleep 1000 & echo -e "Background Process ID=$!" Now, execute the above script, and check the process id which its printing. $ ./proc.sh Process ID=9502 Background Process ID=9503 $ ps PID TTY TIME CMD 5970 pts/1 00:00:00 bash 9503 pts/1 00:00:00 sleep 9504 pts/1 00:00:00 ps $ Example 4: Other Bash Special Parameters – $?, $-, $_ $? Gives the exit status of the most recently executed command. $- Options set using set builtin command $_ Gives the last argument to the previous command. At the shell startup, it gives the absolute filename of the shell script being executed. $ cat others.sh #!/bin/bash echo -e "$_"; ## Absolute name of the file which is being executed /usr/local/bin/dbhome # execute the command. #check the exit status of dbhome if [ "$?" -ne "0" ]; then echo "Sorry, Command execution failed !" fi echo -e "$-"; #Set options - hB echo -e $_ # Last argument of the previous command. In the above script, the last echo statement “echo -e $_” ($ underscore) also prints hB which is the value of last argument of the previous command. So $_ will give the value after expansion $ ./others.sh ./others.sh /home/oracle Sorry, Command execution failed ! hB hB Tweet > Add your comment If you enjoyed this article, you might also like.. 50 Linux Sysadmin Tutorials 50 Most Frequently Used Linux Commands (With Examples) Top 25 Best Linux Perfo
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
android - Select all text inside EditText when it gets focus - Stack Overflow
sked 9 years, 1 month ago Active 20 days ago Viewed 119k times 225 27 [emptylink] I have an EditText with some dummy text in it. When the user clicks on it I want it to be selected so that when <span>the user starts typing the dummy text gets deleted. How can I achieve this? android android-edittext share |improve this question asked Jan 12 '11 at 13:47 [emptylink] [imagelink] Galip Galip 4,91599 gold badges3333 silver badges4747 bro
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Sato_Wider_Windheuser-2019-Continuous_Delivery_Machine_Learning-martinfowler,com
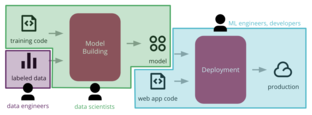
are worried about building and improving the ML model. Then Machine Learning Engineers or developers will have to worry about how to integrate that model and release it to production. Figure 4: <span>common functional silos in large organizations can create barriers, stifling the ability to automate the end-to-end process of deploying ML applications to production This leads to delays and friction. A common symptom is having models that only work in a lab environment and never leave the proof-of-concept phase. Or if they make it to production, in a manual ad-hoc way, they become stale and hard to update. The second challenge is technical: how to make the process reproducible and auditable. Because these teams use different tools and follow different workflows, it becomes hard to automat
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Sato_Wider_Windheuser-2019-Continuous_Delivery_Machine_Learning-martinfowler,com
is having models that only work in a lab environment and never leave the proof-of-concept phase. Or if they make it to production, in a manual ad-hoc way, they become stale and hard to update. <span>The second challenge is technical: how to make the process reproducible and auditable. Because these teams use different tools and follow different workflows, it becomes hard to automate it end-to-end. There are more artifacts to be managed beyond the code, and versioning them is not straightforward. Some of them can be really large, requiring more sophisticated tools to store and ret
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Sato_Wider_Windheuser-2019-Continuous_Delivery_Machine_Learning-martinfowler,com
ess encompasses a major part of a Data Scientist's workflow [1] , for the purposes of CD4ML, we treat the ML pipeline as the final automated implementation of the chosen model training process. <span>Once the data is available, we move into the iterative Data Science workflow of model building. This usually involves splitting the data into a training set and a validation set, trying different combinations of algorithms, and tuning their parameters and hyper-parameters. That produces a model that can be evaluated against the validation set, to assess the quality of its predictions. The step-by-step of this model training process becomes the machine learning pipeline. In Figure 5 we show how we structured the ML pipeline for our sales forecasting problem, highlighting the different source code, data, and model components. The input data, the intermed
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Sato_Wider_Windheuser-2019-Continuous_Delivery_Machine_Learning-martinfowler,com
comes the machine learning pipeline. In Figure 5 we show how we structured the ML pipeline for our sales forecasting problem, highlighting the different source code, data, and model components. <span>The input data, the intermediate training and validation data sets, and the output model can potentially be large files, which we don't want to store in the source control repository. Also, the stages of the pipeline are usually in constant change, which makes it hard to reproduce them outside of the Data Scientist's local environment. Figure 5: Machine Learning pipeline for our Sales Forecasting problem, and the 3 steps to automate it with DVC In order to formalise the model training process in code, we used an open
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Sato_Wider_Windheuser-2019-Continuous_Delivery_Machine_Learning-martinfowler,com
makes it hard to reproduce them outside of the Data Scientist's local environment. Figure 5: Machine Learning pipeline for our Sales Forecasting problem, and the 3 steps to automate it with DVC <span>In order to formalise the model training process in code, we used an open source tool called DVC (Data Science Version Control). It provides similar semantics to Git, but also solves a few ML-specific problems: it has multiple backend plugins to fetch and store large files on an external storage outside of the source control repository; it can keep track of those files' versions, allowing us to retrain our models when the data changes; it keeps track of the dependency graph and commands used to execute the ML pipeline, allowing the process to be reproduced in other environments; it can integrate with Git branches to allow multiple experiments to co-exist; For example, we can configure our initial ML pipeline in Figure 5 with three dvc run commands (-d specify dependencies, -o specify outputs, -f is the filename to record that step, and -
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Sato_Wider_Windheuser-2019-Continuous_Delivery_Machine_Learning-martinfowler,com
cy graph and commands used to execute the ML pipeline, allowing the process to be reproduced in other environments; it can integrate with Git branches to allow multiple experiments to co-exist; <span>For example, we can configure our initial ML pipeline in Figure 5 with three dvc run commands (-d specify dependencies, -o specify outputs, -f is the filename to record that step, and -M is the resulting metrics): dvc run -f input.dvc \ ➊ -d src/download_data.py -o data/raw/store47-2016.csv python src/download_data.py dvc run -f split.dvc \ ➋ -d data/raw/store47-2016.csv -d src/splitter.py \ -o data/splitter/train.csv -o data/splitter/validation.csv python src/splitter.py dvc run ➌ -d data/splitter/train.csv -d data/splitter/validation.csv -d src/decision_tree.py \ -o data/decision_tree/model.pkl -M results/metrics.json python src/decision_tree.py Each run will create a corresponding file, that can be committed to version control, and that allows other people to reproduce the entire ML pipeline, by executing the dvc repro command. Once we find a suitable model, we will treat it as an artifact that needs to be versioned and deployed to production. With DVC, we can use the dvc push and dvc pull commands to publish and fetch it from external storage. There are other open source tools that you can use to solve these problems: Pachyderm uses containers to execute the different steps of the pipeline and also solves the data versioning