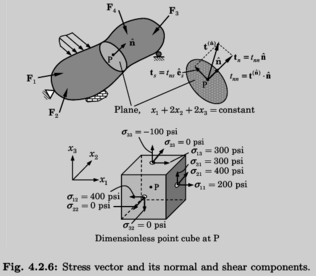
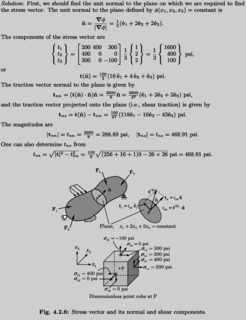
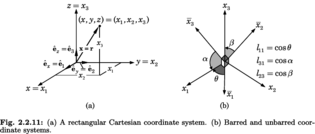
Edited, memorised or added to reading queue
on 03-Mar-2021 (Wed)
Do you want BuboFlash to help you learning these things? Click here to log in or create user.
Flashcard 6180532784396
| status | not learned | measured difficulty | 37% [default] | last interval [days] | |||
|---|---|---|---|---|---|---|---|
| repetition number in this series | 0 | memorised on | scheduled repetition | ||||
| scheduled repetition interval | last repetition or drill |
Flashcard 6180543270156
| status | not learned | measured difficulty | 37% [default] | last interval [days] | |||
|---|---|---|---|---|---|---|---|
| repetition number in this series | 0 | memorised on | scheduled repetition | ||||
| scheduled repetition interval | last repetition or drill |
Flashcard 6180612738316
| status | not learned | measured difficulty | 37% [default] | last interval [days] | |||
|---|---|---|---|---|---|---|---|
| repetition number in this series | 0 | memorised on | scheduled repetition | ||||
| scheduled repetition interval | last repetition or drill |
Flashcard 6217725250828
| status | not learned | measured difficulty | 37% [default] | last interval [days] | |||
|---|---|---|---|---|---|---|---|
| repetition number in this series | 0 | memorised on | scheduled repetition | ||||
| scheduled repetition interval | last repetition or drill |
Flashcard 6217733377292
| status | not learned | measured difficulty | 37% [default] | last interval [days] | |||
|---|---|---|---|---|---|---|---|
| repetition number in this series | 0 | memorised on | scheduled repetition | ||||
| scheduled repetition interval | last repetition or drill |
Flashcard 6220118625548
| status | not learned | measured difficulty | 37% [default] | last interval [days] | |||
|---|---|---|---|---|---|---|---|
| repetition number in this series | 0 | memorised on | scheduled repetition | ||||
| scheduled repetition interval | last repetition or drill |
Emulsão – Wikipédia, a enciclopédia livre
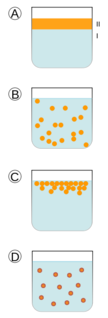
emulsão óleo em água (O/A). Ao contrário, quando a fase interna é aquosa e a externa oleosa, denominamos emulsão água em óleo (A/O)[1]. Estabilidade das emulsões[editar | editar código-fonte] <span>A.. Dois líquidos imiscíveis separados em duas fases (I e II). B. Emulsão da fase II dispersa na fase I. C. A emulsão instável progressivamente retorna ao seu estado inicial de fases separadas. D. O surfactante se posiciona na interface entre as fases I e II, estabilizando a emulsão As emulsões são instáveis termodinamicamente e, portanto não se formam espontaneamente, sendo necessário fornecer energia para formá-las através de agitação, de homogeneizadores, ou de
emulsão óleo em água (O/A). Ao contrário, quando a fase interna é aquosa e a externa oleosa, denominamos emulsão água em óleo (A/O)[1]. Estabilidade das emulsões[editar | editar código-fonte] <span>A.. Dois líquidos imiscíveis separados em duas fases (I e II). B. Emulsão da fase II dispersa na fase I. C. A emulsão instável progressivamente retorna ao seu estado inicial de fases separadas. D. O surfactante se posiciona na interface entre as fases I e II, estabilizando a emulsão As emulsões são instáveis termodinamicamente e, portanto não se formam espontaneamente, sendo necessário fornecer energia para formá-las através de agitação, de homogeneizadores, ou de
Flashcard 6249311767820
| status | not learned | measured difficulty | 37% [default] | last interval [days] | |||
|---|---|---|---|---|---|---|---|
| repetition number in this series | 0 | memorised on | scheduled repetition | ||||
| scheduled repetition interval | last repetition or drill |
Flashcard 6249409809676
| status | not learned | measured difficulty | 37% [default] | last interval [days] | |||
|---|---|---|---|---|---|---|---|
| repetition number in this series | 0 | memorised on | scheduled repetition | ||||
| scheduled repetition interval | last repetition or drill |
Flashcard 6249420557580
| status | not learned | measured difficulty | 37% [default] | last interval [days] | |||
|---|---|---|---|---|---|---|---|
| repetition number in this series | 0 | memorised on | scheduled repetition | ||||
| scheduled repetition interval | last repetition or drill |
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Métabolomique — Wikipédia
lus Modifier les liens Espaces de noms Article Discussion Variantes Affichages Lire Modifier Modifier le code Voir l’historique Plus Métabolomique Un article de Wikipédia, l'encyclopédie libre. <span>La métabolomique est une science très récente qui étudie l'ensemble des métabolites primaires (sucres, acides aminés, acides gras, etc.) et des métabolites secondaires dans le cas des plantes (polyphénols, flavonoïdes, alcaloïdes, etc.) présents dans une cellule, un organe, un organisme. C'est l'équivalent de la génomique pour l'ADN. Elle utilise la spectrométrie de masse et la résonance magnétique nucléaire. Applications[modifier | modifier le code] Médecine : selon d
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
ites The central dogma of biology showing the flow of information from DNA to the phenotype. Associated with each stage is the corresponding systems biology tool, from genomics to metabolomics. <span>Metabolomics is the scientific study of chemical processes involving metabolites, the small molecule substrates, intermediates and products of cell metabolism. Specifically, metabolomics is the "systematic study of the unique chemical fingerprints that specific cellular processes leave behind", the study of their small-molecule metabolite profiles.[1] The metabolome represents the complete set of metabolites in a biological cell, tissue, organ or organism, which are the end products of cellular processes.[2] Messenger RNA (mRNA), gen
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
-molecule metabolite profiles.[1] The metabolome represents the complete set of metabolites in a biological cell, tissue, organ or organism, which are the end products of cellular processes.[2] <span>Messenger RNA (mRNA), gene expression data and proteomic analyses reveal the set of gene products being produced in the cell, data that represents one aspect of cellular function. Conversely, metabolic profiling can give an instantaneous snapshot of the physiology of that cell,[3] and thus, metabolomics provides a direct "functional readout of the physiological state" of an organism.[4] One of the challenges of systems biology and functional genomics is to integrate genomics, transcriptomic, proteomic, and metabolomic information to provide a better understanding of ce
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
omics 6 Analytical technologies 6.1 Separation methods 6.2 Detection methods 7 Statistical methods 8 Key applications 9 See also 10 References 11 Further reading 12 External links History[edit] <span>The concept that individuals might have a "metabolic profile" that could be reflected in the makeup of their biological fluids was introduced by Roger Williams in the late 1940s,[5] who used paper chromatography to suggest characteristic metabolic patterns in urine and saliva were associated with diseases such as schizophrenia. However, it was only through technological advancements in the 1960s and 1970s that it became feasible to quantitatively (as opposed to qualitatively) measure metabolic profiles. [6] Th
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
zophrenia. However, it was only through technological advancements in the 1960s and 1970s that it became feasible to quantitatively (as opposed to qualitatively) measure metabolic profiles. [6] <span>The term "metabolic profile" was introduced by Horning, et al. in 1971 after they demonstrated that gas chromatography-mass spectrometry (GC-MS) could be used to measure compounds present in human urine and tissue extracts.[7][8] The Horning group, along with that of Linus Pauling and Arthur B. Robinson led the development of GC-MS methods to monitor the metabolites present in urine through the 1970s.[9] Concurr
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
[when?] efforts to utilize NMR for metabolomics have been largely driven by the laboratory of Jeremy K. Nicholson at Birkbeck College, University of London and later at Imperial College London. <span>In 1984, Nicholson showed 1H NMR spectroscopy could potentially be used to diagnose diabetes mellitus, and later pioneered the application of pattern recognition methods to NMR spectroscopic data.[12][13] In 1995 liquid chromatography mass spectrometry metabolomics experiments[14] were performed by Gary Siuzdak while working with Richard Lerner (then president of The Scripps Research Ins
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
leamide, was observed and later shown to have sleep inducing properties. This work is one of the earliest such experiments combining liquid chromatography and mass spectrometry in metabolomics. <span>In 2005, the first metabolomics tandem mass spectrometry database, METLIN,[15][16] for characterizing human metabolites was developed in the Siuzdak laboratory at The Scripps Research Institute. METLIN has since grown and as of July 1, 2019, METLIN contains over 450,000 metabolites and other chemical entities, each compound having experimental tandem mass spectrometry data generated from molecular standards at multiple collision energies and in positive and negative ionization modes. METLIN is the largest repository of tandem mass spectrometry data of its kind. 2005 was also the year in which the dedicated academic journal Metabolomics first appeared, founded by its
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
em mass spectrometry data of its kind. 2005 was also the year in which the dedicated academic journal Metabolomics first appeared, founded by its current editor-in-chief Professor Roy Goodacre. <span>In 2005, the Siuzdak lab was engaged in identifying metabolites associated with sepsis and in an effort to address the issue of statistically identifying the most relevant dysregulated metabolites across hundreds of LC/MS datasets, the first algorithm was developed to allow for the nonlinear alignment of mass spectrometry metabolomics data. Called XCMS,[17] where the "X" constitutes any chromatographic technology, it has since (2012)[18] been developed as an online tool and as of 2019 (with METLIN) has over 30,000 registered users. On 23 January 2007, the Human Metabolome Project, led by David Wishart of the University of Alberta, Canada, completed the first draft of the human metabolome, consisting of a database
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
, 1200 drugs and 3500 food components.[19][20] Similar projects have been underway in several plant species, most notably Medicago truncatula[21] and Arabidopsis thaliana[22] for several years. <span>As late as mid-2010, metabolomics was still considered an "emerging field".[23] Further, it was noted that further progress in the field depended in large part, through addressing otherwise "irresolvable technical challenges", by technical evolution of mass spectro
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
ted that further progress in the field depended in large part, through addressing otherwise "irresolvable technical challenges", by technical evolution of mass spectrometry instrumentation.[23] <span>In 2015, real-time metabolome profiling was demonstrated for the first time.[24] Metabolome[edit] Human metabolome project See also: Metabolome and Human Metabolome Database The metabolome refers to the complete set of small-molecule (<1.5 kDa)[25] metabolites (s
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
nstrumentation.[23] In 2015, real-time metabolome profiling was demonstrated for the first time.[24] Metabolome[edit] Human metabolome project See also: Metabolome and Human Metabolome Database <span>The metabolome refers to the complete set of small-molecule (<1.5 kDa)[25] metabolites (such as metabolic intermediates, hormones and other signaling molecules, and secondary metabolites) to be found within a biological sample, such as a single organism.[26][27] The word was coined in analogy with transcriptomics and proteomics; like the transcriptome and the proteome, the metabolome is dynamic, changing from second to second. Although the meta
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
of 2019 (with METLIN) has over 30,000 registered users. In January 2007, scientists at the University of Alberta and the University of Calgary completed the first draft of the human metabolome. <span>The Human Metabolome Database (HMDB) is perhaps the most extensive public metabolomic spectral database to date.[28] The HMDB stores more than 40,000 different metabolite entries. They catalogued approximately 2500 metabolites, 1200 drugs and 3500 food components that can be found in the human body, as reported in the literature.[19] This information, available at the Human Metabolome Database (www.hmdb.ca) and based on analysis of information available in the current scientific literature, is far from complete.[29] In contrast, much more is known about the metabolomes of other organisms. For example, over 50,000 metabolites have been characterized from the plant kingdom, and many thousands of meta
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
sms. For example, over 50,000 metabolites have been characterized from the plant kingdom, and many thousands of metabolites have been identified and/or characterized from single plants.[30][31] <span>Each type of cell and tissue has a unique metabolic ‘fingerprint’ that can elucidate organ or tissue-specific information. Bio-specimens used for metabolomics analysis include but not limit to plasma, serum, urine, saliva, feces, muscle, sweat, exhaled breath and gastrointestinal fluid.[32] The ease of collection facilitates high temporal resolution, and because they are always at dynamic equilibrium with the body, they can describe the host as a whole.[33] Genome can tell
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
rointestinal fluid.[32] The ease of collection facilitates high temporal resolution, and because they are always at dynamic equilibrium with the body, they can describe the host as a whole.[33] <span>Genome can tell what could happen, transcriptome can tell what appears to be happening, proteome can tell what makes it happen and metabolome can tell what has happened and what is happening.[34] Metabolites[edit] Metabolites are the substrates, intermediates and products of metabolism. Within the context of metabolomics, a metabolite is usually defined as any molecule less than
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
could happen, transcriptome can tell what appears to be happening, proteome can tell what makes it happen and metabolome can tell what has happened and what is happening.[34] Metabolites[edit] <span>Metabolites are the substrates, intermediates and products of metabolism. Within the context of metabolomics, a metabolite is usually defined as any molecule less than 1.5 kDa in size.[25] However, there are exceptions to this depending on the sample and detection method. For example, macromolecules such as lipoproteins and albumin are reliably detected in NMR-based metabolomics studies of blood plasma.[35] In plant-based metabolomics, it is common to re
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
proteins and albumin are reliably detected in NMR-based metabolomics studies of blood plasma.[35] In plant-based metabolomics, it is common to refer to "primary" and "secondary" metabolites.[3] <span>A primary metabolite is directly involved in the normal growth, development, and reproduction. A secondary metabolite is not directly involved in those processes, but usually has important ecological function. Examples include antibiotics and pigments.[36] By contrast, in human-based metabolomics, it is more common to describe metabolites as being either endogenous (produced by the host organism) or exogenous.[37] [38]Metabolites of forei
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
abolic reactions, where outputs from one enzymatic chemical reaction are inputs to other chemical reactions. Such systems have been described as hypercycles.[citation needed] Metabonomics[edit] <span>Metabonomics is defined as "the quantitative measurement of the dynamic multiparametric metabolic response of living systems to pathophysiological stimuli or genetic modification". The word origin is from the Greek μεταβολή meaning change and nomos meaning a rule set or set of laws.[40] This approach was pioneered by Jeremy Nicholson at Murdoch University and has been used in toxicology, disease diagnosis and a number of other fields. Historically, the metabonomics app
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
mos meaning a rule set or set of laws.[40] This approach was pioneered by Jeremy Nicholson at Murdoch University and has been used in toxicology, disease diagnosis and a number of other fields. <span>Historically, the metabonomics approach was one of the first methods to apply the scope of systems biology to studies of metabolism.[41][42][43] There has been some disagreement over the exact differences between 'metabolomics' and 'metabonomics'. The difference between the two terms is not related to choice of analytical platfo
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
ent, there is a growing consensus that 'metabolomics' places a greater emphasis on metabolic profiling at a cellular or organ level and is primarily concerned with normal endogenous metabolism. <span>'Metabonomics' extends metabolic profiling to include information about perturbations of metabolism caused by environmental factors (including diet and toxins), disease processes, and the involvement of extragenomic influences, such as gut microflora. This is not a trivial difference; metabolomic studies should, by definition, exclude metabolic contributions from extragenomic sources, because these are external to the system being studied. However, in practice, within the field of human disease research there is still a large degree of overlap in the way both terms are used, and they are often in effect synonymous.[44] Exometabolomics[edit] Main article: Exometabolomics Exometabolomics, or "metabolic footprinting", is the study of extracellular metabolites. It uses many techniques from other subfields
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
dentify associations with disease states and outcomes, determine significant correlations, and characterize metabolic signatures with existing biological knowledge.[47] Separation methods[edit] <span>Initially, analytes in a metabolomic sample comprise a highly complex mixture. This complex mixture can be simplified prior to detection by separating some analytes from others. Separation achieves various goals: analytes which cannot be resolved by the detector may be separated in this step; in MS analysis ion suppression is reduced; the retention time of the analyte serves as information regarding its identity. This separation step is not mandatory and is often omitted in NMR and "shotgun" based approaches such as shotgun lipidomics. Gas chromatography (GC), especially when interfaced with mass spectrometry (GC-MS), is a widely used separation technique for metabolomic analysis.[48] GC offers very high chromatograph
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
etabolite classes than is GC. As for all electrophoretic techniques, it is most appropriate for charged analytes.[51] Detection methods[edit] Comparison of most common used metabolomics methods <span>Mass spectrometry (MS) is used to identify and quantify metabolites after optional separation by GC, HPLC, or CE. GC-MS was the first hyphenated technique to be developed. Identification leverages the distinct patterns in which analytes fragment which can be thought of as a mass spectral fingerprint; libraries exist that allow identification of a metabol
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
ESI) is a promising approach to circumvent this limitation.[citation needed] Most recently, ion trap techniques such as orbitrap mass spectrometry are also applied to metabolomics research.[54] <span>Nuclear magnetic resonance (NMR) spectroscopy is the only detection technique which does not rely on separation of the analytes, and the sample can thus be recovered for further analyses. All kinds of small molecule metabolites can be measured simultaneously - in this sense, NMR is close to being a universal detector. The main advantages of NMR are high analytical reproducibility and simplicity of sample preparation. Practically, however, it is relatively insensitive compared to mass spectrometry-bas
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
s, and the sample can thus be recovered for further analyses. All kinds of small molecule metabolites can be measured simultaneously - in this sense, NMR is close to being a universal detector. <span>The main advantages of NMR are high analytical reproducibility and simplicity of sample preparation. Practically, however, it is relatively insensitive compared to mass spectrometry-based techniques.[55][56] Comparison of most common used metabolomics methods is shown in the table. Although NMR and MS are the most widely used, modern day techniques other methods of detection that have been used. These include Fourier-transform ion cyclotron resonance,[57] ion-mobi
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
detection (coupled to HPLC), Raman spectroscopy and radiolabel (when combined with thin-layer chromatography).[citation needed] Statistical methods[edit] Software List for Metabolomic Analysis <span>The data generated in metabolomics usually consist of measurements performed on subjects under various conditions. These measurements may be digitized spectra, or a list of metabolite features. In its simplest form this generates a matrix with rows corresponding to subjects and columns corresponding with metabolite features (or vice versa).[7] Several statistical programs are currently available for analysis of both NMR and mass spectrometry data. A great number of free software are already available for the analysis of metab
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
ions of a dataset to a few which explain the greatest variation.[33] When analyzed in the lower-dimensional PCA space, clustering of samples with similar metabolic fingerprints can be detected. <span>PCA algorithms aim to replace all correlated variables by a much smaller number of uncorrelated variables (referred to as principal components (PCs)) and retain most of the information in the original dataset.[61] This clustering can elucidate patterns and assist in the determination of disease biomarkers - metabolites that correlate most with class membership. Linear models are commonly used for metabolomics data, but are affected by multicollinearity. On the other hand, multivariate statistics are thriving methods for high-dimensional correl
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
xicity and some disease models, the metabolites of interest are not known a priori. This makes unsupervised methods, those with no prior assumptions of class membership, a popular first choice. <span>The most common of these methods includes principal component analysis (PCA) which can efficiently reduce the dimensions of a dataset to a few which explain the greatest variation.[33] When analyzed in the lower-dimensional PCA space, clustering of samples with similar metabolic fingerprints can be detected. PCA algorithms aim to replace all correlated variables by a
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
the toxicity of potential drug candidates: if a compound can be eliminated before it reaches clinical trials on the grounds of adverse toxicity, it saves the enormous expense of the trials.[44] <span>For functional genomics, metabolomics can be an excellent tool for determining the phenotype caused by a genetic manipulation, such as gene deletion or insertion. Sometimes this can be a sufficient goal in itself—for instance, to detect any phenotypic changes in a genetically modified plant intended for human or animal consumption. More exciting
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Metabolomics - Wikipedia
ntion time prediction increases the identification rate in liquid chromatography and subsequently leads to an improved biological interpretation of metabolomics data.[63] Key applications[edit] <span>Toxicity assessment/toxicology by metabolic profiling (especially of urine or blood plasma samples) detects the physiological changes caused by toxic insult of a chemical (or mixture of chemicals). In many cases, the observed changes can be related to specific syndromes, e.g. a specific lesion in liver or kidney. This is of particular relevance to pharmaceutical companies wanting to test the toxicity of potential drug candidates: if a compound can be eliminated before it reaches clinical trials
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
| status | not read | reprioritisations | ||
|---|---|---|---|---|
| last reprioritisation on | suggested re-reading day | |||
| started reading on | finished reading on |
Flashcard 6263381036300
| status | not learned | measured difficulty | 37% [default] | last interval [days] | |||
|---|---|---|---|---|---|---|---|
| repetition number in this series | 0 | memorised on | scheduled repetition | ||||
| scheduled repetition interval | last repetition or drill |